|
|||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||
|
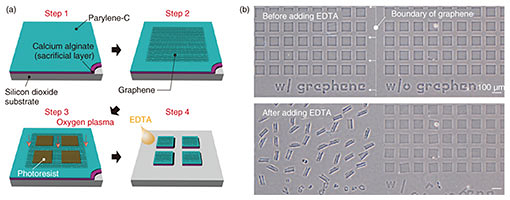
Regular Articles Vol. 18, No. 2, pp. 32–39, Feb. 2020. https://doi.org/10.53829/ntr202002ra1 Self-folded Three-dimensional Graphene for BiointerfacesAbstractThree-dimensional (3D) graphene-based electrodes have been gaining much interest regarding applications in flexible electronics and biointerfaces. We propose a simple method of transforming two-dimensional (2D) monolayer graphene into 3D structures that interface with biological samples. We found that the transferred monolayer graphene tightly adheres to the polymer surface via π-π stacking forces, resulting in the spontaneous folding of graphene/polymer bilayers (self-folding). Owing to the high biocompatibility of the materials and self-folding procedures, the self-folded bilayer films provide a hollow and non-toxic environment to encapsulate and culture cells. The cells embedded in the rolled-up architectures spontaneously form cellular 3D constructs with the intrinsic morphologies and functions of living tissues. For instance, we demonstrate that neuronal constructs with 3D geometry contribute to the construction of brain-like functional tissues such as functional integration with surrounding neuronal networks via their extended axons. Our method can be used to potentially provide 3D biointerfaces necessary for the reconstruction and assembly of functional tissues and implantable tissue grafts. Keywords: self-folding, graphene, neural engineering, tissue regeneration 1. IntroductionGiven its non-cytotoxicity, electrical/thermal conductivity, and mechanical strength, graphene is one of the most promising two-dimensional (2D) materials for applications in flexible bioelectronics [1]. Specifically, its large open-surface enables ions, molecules, and cells to be anchored and desorbed, showing its relevance to electro-chemical sensing [2]. For practical application, the inherently planar geometries of graphene are curved, folded, and wrinkled to assemble three-dimensional (3D) structures and interface with 3D objects. Conventionally, these 3D geometries of graphene have been assembled by loading on flexible templates and applying external mechanical forces [3]. However, this manual process results in technical difficulties in constructing dimensionally well-controlled 3D structures. In addition, the relatively poor adhesion of the transferred graphene leads to detachment and delamination from the substrates, resulting in difficulty in creating 3D graphene architectures. We propose a method for rapid and easy formation of microscopic 3D graphene structures by using origami-inspired self-rolling bilayer films [4]. We use poly(p-xylylene) (parylene) as a flexible template that tightly adheres to graphene due to π-π stacking sp2 hybridization. We discover the self-assembly principle in which the graphene behaves as the driving force behind the transformation of parylene films into tubular and spherical shapes termed a micro-rolls. Simply transferred monolayer graphene forms graphene-laden bilayer films with heterogeneous mechanical properties. Consequently, the differential strain gradients inside the films trigger spontaneous transformation into micro-rolls. To demonstrate the biocompatible 3D graphene interfaces, we used this graphene-laden self-folded cylindrical structure as an interface with neurons [5]. The self-folding graphene film encapsulates neurons inside its folded cylindrical structure, reconstitutes the intrinsic cellular morphologies and functions, and forms functional connections with surrounding neurons. This is potentially applicable to creating 3D bioelectrodes and biointerfaces for applications such as the reconstruction of functional tissues and implantable tissue grafts. 2. Materials and experimental methods2.1 Preparation of materialsOwing to tight π-π adhesion with graphene, we used poly(chloro-p-xylylene) (parylene-C) as a mechanically stable, biocompatible, and aromatic ring-rich polymer [6–8]. We adhered the multi-layered films composed of parylene, polycrystalline / single-crystal monolayer graphene, to the underlying sacrificial layer. While polycrystalline monolayer graphene chemical vapor deposition (CVD)-grown on copper (Cu) foils was purchased from Graphene Platform Corp., large-scale single-crystal graphene was synthesized on the Cu foils with an atmospheric pressure CVD process by using a mixture gas of methane (CH4) and hydrogen (H2) [9]. The sacrificial layer consists of calcium (Ca) alginate hydrogel that is dissolved instantly by adding chelating agents such as ethylenediaminetetraacetic acid (EDTA) or sodium citrate solution. To examine the biocompatibility of self-folded structures, we used primary hippocampal neurons that were dissociated from the hippocampi of Wistar rat embryos. 2.2 Device fabrication and cell encapsulationFigure 1(a) shows the process used to fabricate micro-patterned graphene/parylene bilayers. After Ca alginate was spin-coated on silicon dioxide (SiO2) substrates, both single-crystalline and polycrystalline monolayer graphene were transferred using the conventional poly(methyl methacrylate)-assisted method. Subsequently, parylene-C was deposited using a CVD process and covered with photolithographically micro-patterned photoresist. The triple-layered film was finally etched with oxygen plasma to create a micro-patterned film array. After suspending the neurons, an array of micro-patterned films was released from the substrate by dissolving Ca-alginate sacrificial layers via immersion in EDTA solution. 3. Results3.1 Self-folding of 3D graphene-polymer architecturesThe dissolution of the sacrificial Ca-alginate layer with EDTA resulted in simultaneous batch self-folding of the graphene-laden parylene bilayers into cylindrical micro-rolls (Fig. 1(b)). Interestingly, the bilayer was bent in the direction of the substrates while maintaining the bottom parylene on the inward side. At this time, the loaded graphene appeared to be exposed to tensile force; thus, causing it to cover the inner parylene from the outside. We confirmed that the curvature of the micro-rolls is controllable with the thicknesses of parylene (tp) and the number of graphene layers (tg). Given the transferred monolayer graphene, a thinner tp leads to a smaller micro-roll; stacking multi-layered graphene makes the micro-rolls much tighter when tp is constant. Both reducing the thickness of the parylene layer and sequentially stacking multi-layered graphene decreases the bending rigidity, thereby improving machining controllability of a fine 3D structure. These results indicate that the underlying mechanism of self-folding is the stiffness mismatch in the bilayer, following the trend predicted by the bimorph beam theory [10].
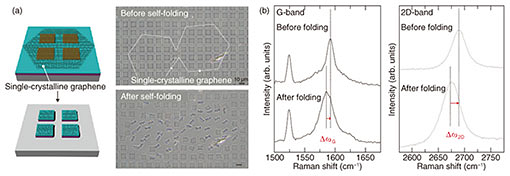
While the self-folding orientation of polycrystalline graphene-laden bilayers was random, one of the single-crystalline graphene-laden bilayers was relatively uniform. Figure 2(a) shows that two adjacent hexagonally shaped-single graphene domains caused the subsequent unidirectional self-folding to form a tubular shape with a uniform curvature. This difference is attributed to a patchwork of relatively smaller single-crystalline grains separated by grain boundaries within the polycrystalline graphene. Since one might expect a single domain of graphene to have the same elastic energy, which induces an isotropic tensile force for folding, the uncontrollable folding orientation originates from domains inside the CVD-grown graphene [11]. In contrast, when the domain size is larger than one sheet of micro-patterned film, the crystalline orientation of graphene would certainly affect the orientation of self-folding. 3.2 Electrical propertiesThe strain in outer graphene during self-folding was estimated by characterizing shifts in G peaks (¦¤G) and 2D peaks (¦¤2D) from the original modes with Raman spectroscopy (Fig. 2(b)). The blue-shifts in both ¦¤G (~8 cm−1) and ¦¤G (~16 cm−1) stem from the distorted graphene lattice and altered interatomic distance, which determines the strain of 0.18%. This estimated strain within the graphene is almost in agreement with the predicted value in the 0.16–0.18% range that is theoretically rationalized with the bimorph-beam theory [10]. Therefore, graphene loaded on parylene will not only longitudinally elongate but also contract transversely with uniaxial tension.
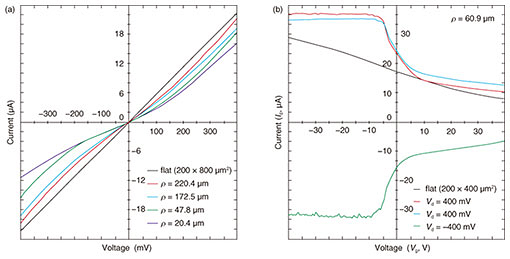
The structural alteration of hexagonal-shaped graphene is critical in terms of changing the electronic structure. Therefore, we newly micro-patterned bilayers that were hinged and partially pinned down and measured their drain current (Id)–voltage (Vd) characteristics before and after self-folding at room temperature. While flat graphene exhibits linear behavior, the self-folding process greatly alters the electrical property of loaded graphene to non-linear (Fig. 3(a)). The resistance increased from 17.9 to 38.0 k¦¸ when measured at 0 mV. When applying back-gate voltages (Vg), both positive and negative back-gate voltages modulated the non-linear resistance of micro-rolls (Fig. 3(b)). This result indicates that the self-folding process never leads to the exfoliation or rupture of graphene and behaviors as a p-type semiconductor in both flat and folded states. We attribute the altered charge carrier densities even at room temperature to the uniformly compressed graphene-unit hexagonal cell. This modulatable property makes it possible to develop switching transistors.
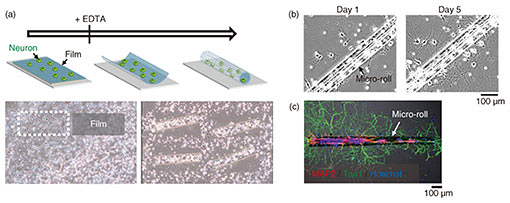
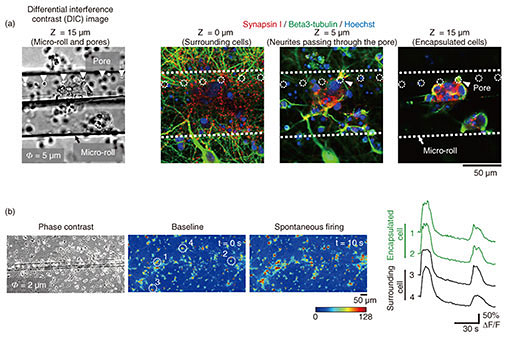
3.3 Encapsulation of cells in porous micro-rollsTo apply graphene micro-rolls to biointerfaces, we investigated their biocompatibility by encapsulating the primary hippocampal neurons inside them [5]. After exposure to EDTA, the cells on the 2D film surface were encapsulated inside the micro-rolls through the self-folding of the bilayer (Fig. 4(a)). Both self-folding and encapsulation were sufficiently gentle to avoid cell damage, which allows the encapsulated cells to migrate inside the micro-rolls during incubation. Since the curvature radius and length of the micro-rolls are controllable, the number of encapsulated cells can be easily controlled with the cell density and volume of the micro-rolls. The high biocompatibility and transparency of the micro-rolls enable stable culturing of encapsulated cells. To enable long-term cell culture, we incorporated the tiny pores with several different diameters to facilitate reagent delivery to the encapsulated cells. Importantly, these numerous pores inside the bilayer have no effect on the curvature radius of the micro-rolls. The pores have two additional characteristics to enable long-term cell culture. First, they allow the neurites to grow across them and extend outside the micro-roll. Time-lapse images of neuron-laden micro-rolls over 5 days show that the neurites from a porous micro-roll protruded beyond the micro-roll 2 h after folding and became longer with an average growth rate of approximately 3 µm/h (Fig. 4(b)). Second, they enable us to load biological probes and pharmacologically stimulate the encapsulated cells. Although a major advantage of cell encapsulation is homogenous cell distribution, long-term culture causes changes in the distribution depending on the cell type and its adhesive and migration properties. The results indicate that porous micro-rolls help encapsulated cells maintain their positions with homogeneous distribution. To further investigate the morphology, the neuron-laden micro-rolls were characterized with immunocytochemistry by using two neuronal markers: MAP2 (dendrite/cell body marker; red) and tau1 (axon/cell body marker; green). Figure 4(c) shows that MAP2-positive neuronal cell bodies were sustainably located in porous micro-rolls and greatly extended their axons (tau1-positive) around the micro-rolls (Φ = 3 µm). We verified that porous self-folding film enables us to isolate axons from neuronal cell bodies cultured in a 3D tubular structure. It is well recognized that a 3D tubular structure is useful for guiding neurite outgrowth. When neurons are encapsulated, the confined structure not only arranges neurons but also guides axons in the single direction as the long axis. Also, incorporated pores on the surface allow both control over the cell distribution and axon guidance in multiple directions. Thanks to the axon guidance in multiple directions, a porous neuron-laden micro-roll can potentially be used as a building block for constructing a complex neuronal network and creating a brain-like network.
3.4 Reconstruction of functional tissue-like structuresThe key challenge with cell-laden micro-rolls is to fabricate tissue-mimicking tubular structures and confirm their cellular function [12]. To demonstrate that pores provide functional integration of neuron-laden micro-rolls into a surrounding neuronal network, a culture was labelled with anti-beta3-tubulin (neuronal marker; green) and anti-synapsin I (synapse marker; red) to assess the morphology and synapse formations around the pores. Confocal microscopic images indicate that beta3-tubulin-positive cells inside and outside the micro-roll were connected by neurites that pass through a pore (Fig. 5(a)). Furthermore, synapsin I was expressed in the neurons, forming synapse puncta along their neurites. Importantly, synapsin I puncta were detected along the neurites passing through the pores (white arrowhead), suggesting the formation of synaptic connections between encapsulated and surrounding neurons. To determine whether a functional synaptic connection is formed, we investigated the synchronization of spontaneous activities within the neuronal network by monitoring intracellular calcium ions (Ca2+). Figure 5(b) shows that spontaneous Ca2+ increases in the neurons attached within and around the micro-rolls. The spontaneous Ca2+ oscillations are highly synchronized between encapsulated and surrounding neurons. Given that synchronized spontaneous activity is typically mediated by glutamate synaptic transmissions in the hippocampal neuronal network, the synchronized Ca2+ oscillations indicate the formation of functional synaptic connections. Hence, the encapsulated neurons were functionally integrated into surrounding neuronal networks by extending their axons through the pores.
It is generally believed that the functional integration of transplanted neurons into existing circuitry is required for neural transplantation leading to brain repair. Although cell encapsulation is a useful method for 3D tissue formation and cell delivery, the confined structure limits the formation of neuronal connections with surrounding tissues. Cell encapsulation using a porous film allows encapsulated neurons to extend their axons across the pores and form functional synaptic connections with surrounding neurons. These results highlight the potential for using a neuron-laden micro-roll with pores as a platform for constructing functional neural tissue with highly precise 3D geometry. Furthermore, the graphene surrounding a micro-roll will be used as an electrode for electrophysiological recording and the stimulation of neuronal activity. 4. SummaryWe proposed a simple method of transforming a monolayer graphene-laden polymeric film (parylene) into a programmed 3D configuration that depends on 2D geometry. The adhesion between graphene and parylene is mainly attributable to intermolecular force including π-π stacking, which ensures a conformal and stable contact within bilayers. The strain gradient in homogeneous films initiates self-folding into micro-rolls. Remarkably, micro-scale curved graphene exhibits previously unachievable non-linear electrical behavior without any fracturing or insulation. In addition, both the fabrication process and materials of micro-rolls never require cytotoxic etchant, allowing for batch encapsulation of multiple cells. The biocompatibility of graphene and parylene also ensure the encapsulation and long-term culturing of more than one month of encapsulated cells without cytotoxicity. Notably, the higher elasticity of graphene and parylene enables the embedded cell to develop a fiber-shaped construct. In particular, a porous graphene/parylene bilayer can encapsulate neurons and allow these neurons to interact with their surroundings. The pores facilitate the diffusion of reagents to encapsulated neurons for the dye loading and stimulation essential for the analysis of neuronal circuits. As a result, the encapsulated neurons can be functionally integrated into surrounding neuronal networks by extending their axons through the pores. Thus, the porous self-folding film allows us to construct neuronal tissues connecting to surrounding tissues with a precisely controlled cell distribution. This method could be expanded for use with many other adherent cell lines for reconstructing fiber-shaped functional tissues that mimic muscle fibers, blood vessels, and nerve networks in vitro. 5. Future prospectsFrom the perspective of material science, in combination with the 3D reconfigurable structure with electrical conductivity, this method could be extensively used for the multi-functional integrated circuits needed for flexible electronics and field effect devices in which the electrical conductivity of graphene is tunable with self-folded geometries. Furthermore, the ability to fold the atomically thin sheet into 3D shapes allows the formation of multi-functional micro-rolls with diverse chemical, electrical, spintronic, and optical characteristics. From the biological perspectives, the method can also be used to form scaffolding structures toward regenerative medicine and the behavior analysis of single cells. This method could be expanded to reconstruct tissues with various shapes in vitro that are suitable for exploring single-cell assays, tissue engineering, and implantable ex vivo tissue grafts. In addition, a graphene electrode can be implanted in the brain surface for long-term electrophysiological recording. Therefore, if graphene can be attached to both the inner and outer face, it will be possible to record neuronal activities in encapsulated and surrounding tissue independently and investigate the establishment of functional connections. We believe that the incorporation of the electrical functionality of graphene will have great potential in terms of developing an integrated platform for both cell transplantation and electrophysiological recording. AcknowledgmentsWe are grateful to Dr. Makoto Takamura, Dr. Yui Ogawa, and Dr. Shengnan Wang for providing monolayer CVD graphene samples and fruitful discussions regarding graphene characterization. We are also grateful to Dr. Yoshiaki Kashimura and Yuriko Furukawa for their help with the microscopic observation, characterization of electrical property, and assistance regarding cell culture. This work was partially supported by Grants-in-Aid for Scientific Research (JP17H02759) from the Japan Society for the Promotion of Science. References
|
|||||||||||||||||||||||||