|
|||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||
|
Regular Articles Vol. 21, No. 6, pp. 58–65, June 2023. https://doi.org/10.53829/ntr202306ra1 Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in a Unicellular Red AlgaAbstractTriacylglycerols (TAGs) generated by microalgae are a raw material for liquid biofuel production, so increasing the amount of TAGs generated will contribute to reducing the environmental impact of, for example, greenhouse gas emissions. Since transcription factors (TFs) regulate the expression of a group of genes with related functions, it is thought that TAG accumulation can be enhanced by identifying TFs involved in TAG accumulation and enhancing their functionality. In this study, my research colleagues and I used transcriptomic and phosphoproteomic data—obtained under conditions of TAG accumulation in a unicellular red alga, Cyanidioschyzon merolae, to identify 14 TFs that may regulate TAG accumulation. To verify the function of these TFs, we constructed functionally enhanced strains overexpressing each TF and analyzed changes in TAG accumulation. The analysis results indicate that the amount of TAGs regarding the four overexpressing strains increased 2.2 to 3.8 times compared with that regarding the control strain, so we can consider that those four TFs are involved in TAG accumulation. Transcriptome analysis of each of the four TF overexpression strains showed that among the group of genes related to TAG synthesis, only a gene-encoding endoplasmic reticulum-localized lysophosphatidic acid acyltransferase 1 (LPAT1) significantly enhanced. In strains that overexpressed LPAT1 and enhanced its functionality, TAG accumulation increased 3.3 times compared with that in the control strain. These results (i) explain the mechanism by which four types of TFs regulate the TAG amount in C. merolae by altering the expression of a group of target genes, including LPAT1, and (ii) indicate that enhancing TAG accumulation by strengthening functionality TF is useful. Keywords: algal biofuel, lysophosphatidic acid acyltransferase, red alga, transcription factor, triacylglycerol 1. IntroductionMicroalgae store excess energy in lipid droplets (LDs), which are mainly composed of neutral lipids, in their cytoplasm. Neutral lipids contained in LDs are mainly triacylglycerols (TAGs), which can be used as a raw material for producing biofuel. Since the carbon that constitutes TAGs is derived from carbon dioxide (CO2) fixed by photosynthesis, biofuel production using microalgae is expected to contribute to reducing environmental load such as mitigating global warming [1–3]. However, a large-scale system for industrial biofuel production using microalgae has not been established. The main reason is due to the lack in the understanding of the underlying molecular mechanisms that control TAG accumulation in microalgae. Our research group has aimed to explain the regulatory mechanism of TAG accumulation by using a model unicellular red alga Cyanidioschyzon merolae [4]. It has been observed that exposure of C. merolae to nitrogen-deficient (-N) conditions enhances TAG accumulation [5], which has been observed for many other microalgae. The target of rapamycin (TOR) has also been shown to govern the accumulation of TAGs under -N conditions [5]. TOR is a highly conserved protein kinase among eukaryotes and plays a central role in cell growth and stress response by sensing environmental conditions [6]. The kinase activity of TOR is specifically inhibited by rapamycin, an inhibitor of TOR [7]. In our previous studies, when rapamycin-sensitive C. merolae strains F12 or SF12 [8, 9] were grown under normal culture conditions in which TAGs do not accumulate, the addition of rapamycin to the culture medium resulted in accumulation of almost the same amount of TAGs as observed under -N conditions [5, 9]. TAG accumulation after inhibition of TOR activity has also been observed in other algae [10–12], in other words, the mechanism that regulates TAG accumulation by TOR has been shown to be common to all algae [13]. Several studies have reported on the use of over expression of a single gene involved in synthesis of TAGs as a method for enhancing TAG accumulation [14, 15] (Fig. 1). Compared with such methods, simultaneously enhancing the expression of multiple genes involved in TAG synthesis could further enhance TAG accumulation. One way to achieve this simultaneous multiple-gene enhancement is to enhance the function of the transcription factors (TFs) that control gene expression. This is possible because each TF regulates a group of genes with related functions [16] (Fig. 1).
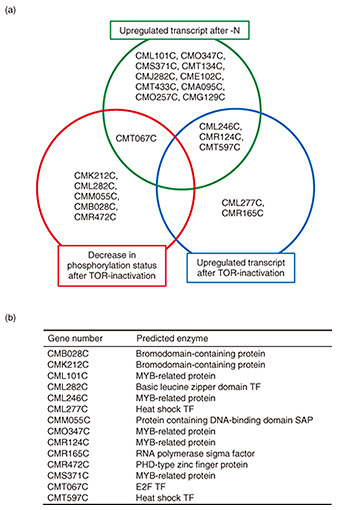
The aims of this study were (i) identify TFs involved in TAG accumulation in C. merolae, (ii) enhance TAG accumulation by strengthening the functions of the TFs, and (iii) identify genes that are regulated by the identified TFs. 2. Identification of candidate TFs involved in TAG accumulationUsing previously reported microarray data [5, 10] and phosphoproteome data [17] obtained under -N- and TOR-activity-inhibiting conditions (under which TAG accumulation in C. merolae is enhanced), we identified genes encoding TF with an increase in transcripts and TFs with varying degrees of phosphorylation. The TFs identified for each condition are shown in the Venn diagram in Fig. 2(a). The notation of each TF corresponds to the number in the C. merolae database (http://czon.jp). For the ten TFs with expression enhanced only under -N conditions, the number of TF candidates was narrowed due to their large number. In green algae, ROC40 (an MYB-type TF) has been reported to be involved in TAG accumulation under -N conditions [18]. Accordingly, among the ten candidate TFs, the ROC40 homolog CML101C, CMO347C, and CMS371C were analyzed. A total of 14 types of TFs (as listed in Fig. 2(b)) were therefore analyzed. Note that the four TFs, CML246C, CMR124C, CMT597C, and CMT067C, showed enhancement under two conditions.
3. Enhancement of LD formation and TAG accumulation by strengthening the function of TFsIf the 14 candidate TFs play critical roles in TAG accumulation in C. merolae, overexpression of each TF would potentially lead to an accumulation of cytoplasmic LDs that contain TAGs. To examine this possibility, we constructed overexpression strains for each candidate TF. It should be noted that the overexpression strain for CMT597C has not yet been successfully constructed. The reason is unknown, but it is conceivable that enhanced expression of the CMT597C protein may lead to cell death. Accordingly, CMT597C was excluded from subsequent analysis, leaving a total of 13 TFs included in the analysis (Fig. 3). The name of each overexpression strain is shown with “ox” after the gene number. For example, for CMB028C, the name of its overexpression strain is CMB028Cox.
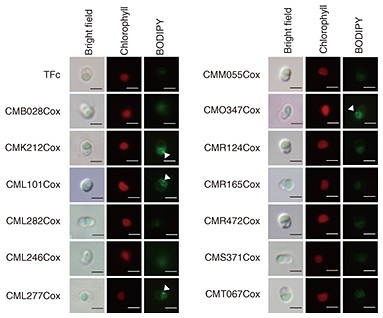
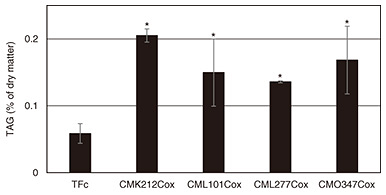
Each of the generated overexpression strains was grown under normal culture conditions under which TAGs do not accumulate as the wild type, and the presence or absence of LD formation was determined. LDs were stained with BODIPY (4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene), i.e., a fluorescent reagent for neutral lipids, and observed using a fluorescence microscope. The observation results indicate clear LD formation in the CMK212Cox, CML101Cox, CML277Cox, and CMO347Cox strains (Fig. 3, white arrowheads). Chlorophyll indicates the autofluorescence of chloroplasts, namely, the localization of chloroplasts, and scale bars indicate 2 μm. TAG accumulation in each strain was then measured using gas chromatography under the same conditions as when LD formation was observed. The results indicate that TAG accumulation in the CMK212Cox, CML101Cox, CML277Cox, and CMO347Cox strains significantly enhanced—2.2, 3.2, 3.8, and 2.7 times, respectively—compared with the control strain TFc (Fig. 4). These results indicate that these four TFs function positively in regard to TAG accumulation in C. merolae.
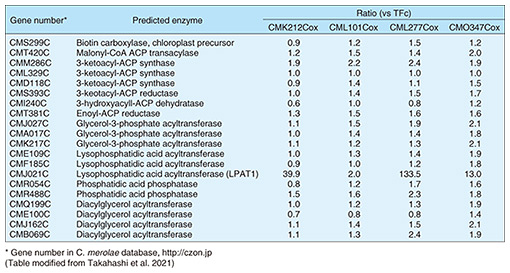
4. Comparison of expression levels of TAG and fatty-acid synthesis-related genes in CMK212Cox, CML101Cox, CML277Cox, and CMO347Cox strainsThe results described in the previous section indicate that the increase in TAG accumulation due to overexpression of the four TFs strains (CMK212Cox, CML101Cox, CML277Cox, and CMO347Cox) is caused by changes in the expression of genes regulated by those TFs, particularly a group of genes involved in TAG synthesis. Therefore, we conducted transcriptome analysis (using a next-generation sequencer) on ribonucleic acid (RNA) isolated from cells of each overexpression strain and control strain TFc under normal culture conditions. Table 1 shows the variation in gene-expression ratios of the gene groups involved in synthesis of TAGs and synthesis of fatty acids, which are materials for TAG synthesis, with the relative value of TFc taken as 1. As shown in Table 1, only CMJ021C, which encodes lysophosphatidic acid acyltransferase (labelled “LPAT1” on the table), showed enhanced expression for all four overexpression strains, i.e., expression ratio increased (compared with that of TFc) 39.9 times for CMK212Cox, 2.0 times for CML101Cox, 133.5 times for CML277Cox, and 13.0 times for CMO347Cox. These results suggest that the TAG accumulation observed in the four overexpression strains may be due to increased expression of LPAT1.
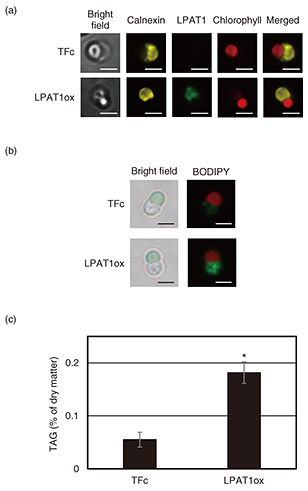
5. Enhancement of LD formation and TAG accumulation by enhancing functionality of LPAT1To investigate the role of C. merolae LPAT1 in TAG synthesis and its subcellular localization, we generated an overexpression strain of LPAT1, i.e., LPAT1ox. In accordance with the results of the immunostaining analysis shown in Fig. 5(a), a signal derived from LPAT1 (green in the figure) was detected at the same location as calnexin (yellow in the figure) localized in the endoplasmic reticulum, which is the site of TAG synthesis. Chlorophyll (red in the figure) indicates chloroplast autofluorescence and chloroplast localization, and “Merged” is a superimposed image of the calnexin, LPAT1, and chlorophyll images in which the scale bar indicates 2 μm. In accordance with the results of the analysis of LD formation and TAG accumulation in the LPAT1ox strain shown in Figs. 5(b) and (c), respectively, LD formation enhanced, and TAG accumulation increased significantly (3.3 times compared with that for TFc). These results indicate that the reaction catalyzed by LPAT1, which localizes to the endoplasmic reticulum, is the rate-limiting step of TAG synthesis in C. merolae and is a key factor in TAG accumulation (Fig. 6).
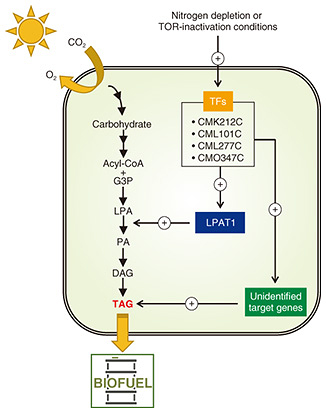
6. Conclusion and future directionsAnalysis based on transcriptomic and phosphoproteomic data identified four TFs involved in TAG accumulation. No other large-scale analysis has identified multiple TFs involved in TAG accumulation in a single algal species. Moreover, the results of analyzing those target genes revealed that (i) LPAT1 localized in the endoplasmic reticulum and (ii) enhancement of its function enhanced accumulation of TAGs. On the basis of the above results, Fig. 6 shows a possible model in which G3P, LPA, PA, DAG, and TAG respectively stand for glycerol-3-phosphate, lysophosphatidic acid, phosphatidic acid, diacylglycerol, and triacylglycerol, and the circled + indicates a positive effect. Conditions under which TAG accumulates, such as -N conditions or inhibition of TOR activity, the expression of four TFs, CMK212C, CML101C, CML277C, and CMO347C, is enhanced, and subsequent upregulation of LPAT1 of their target genes leads to TAG accumulation [19]. Target genes other than LPAT1 of the four TFs (labelled “Unidentified target genes” in Fig. 6) are unknown at this time and should be further analyzed. Simultaneous enhancement of these genes (after they are revealed) in addition to the function of LPAT1 is expected to further enhance TAG accumulation. It is thus concluded that identification of TFs involved in TAG accumulation is an excellent strategy for increasing TAG content and will provide important information for biofuel production using microalgae. References
|
|||||||||||||||||||||||||||||||||||||||