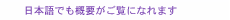|
|||||||||||||||||||||||
|
|
|||||||||||||||||||||||
|
Special Feature: Nanobiotechnology Research Vol. 7, No. 8, pp. 15–19, Aug. 2009. https://doi.org/10.53829/ntr200908sf3 Direct Observation of Dynamics in Receptor Protein by Atomic Force MicroscopyAbstractThis article describes recent results that show the structure of and conformational changes in a single receptor protein observed by atomic force microscopy. A receptor is a functional protein present in our bodies, and it has various physiological functions and is affected in various ways by drugs. This new understanding of receptor function and structure will help to clarify how receptors perform their physiological functions and will lead to the fabrication of new functional devices that combine biological molecules and artificial materials.
1. IntroductionA receptor is the means by which cells receive extracellular information. In most cases, receptors are membrane proteins that exist in cell membranes, and they account for about one third of the one hundred thousand types of human proteins. Receptors function by binding with biologically active agents, including hormones and drugs. An under- or overactive receptor function contributes to the development of various pathological conditions. Drug treatment improves the pathology by regulating the receptor function. A recent survey has revealed that receptors constitute about 45% of drug targets [1]. Therefore, receptors are an important area of study in drug development. In the brain, many kinds of compounds such as peptides, amino acids (e.g., glutamate and γ-amino butyric acid), hormones (e.g., catecholamine), and others (e.g., adenosine nucleotides) function as intercellular transmitters (neurotransmitters) that bind to receptors in cell membranes. Receptors are divided into two families: G protein-coupled receptors (GPCR, where G stands for guanine nucleotide-binding protein), which are metabotropic receptors, and ionotropic receptors. With GPCR, when a receptor is bound to its ligand, it converts intracellular G protein from a guanosine-diphosphate-bound type to a guanosine-triphosphate-bound type, thereby transducing signals. On the other hand, the structures of ionotropic receptors change in response to ligand binding in their extracellular domain, resulting in the opening of a transmembrane pore. Ions, such as Na+, and Ca2+, flow through this pore (Fig. 1), and this flow is recognized as a signal by the cell. In this article, we discuss results [2] that we obtained for the direct imaging of a single ionotropic receptor by atomic force microscopy (AFM). Most ionotropic receptors form a functional channel by assembling several receptor proteins. For example, as we describe in this article, the structures of three adenosine triphosphate (ATP) receptors combine to form one functional ion channel (ATP receptor channel). As described above, pore opening is required to allow ion channels to perform their function, so it is important to analyze their structure in order to understand the receptor function mechanism. X-ray diffraction is a well-known method that can analyze protein structure with high spatial resolution, but it requires crystallized proteins. The crystallized condition is different from the in-vivo condition, and the structural dynamics in crystallized proteins are difficult to observe. To address this issue, we used AFM to observe proteins with high spatial resolution in the time domain in liquid. AFM is a well-known method that detects the topology of a sample. It can be used for biological molecules including deoxyribonucleic acid (DNA), lipids, and proteins in liquid.
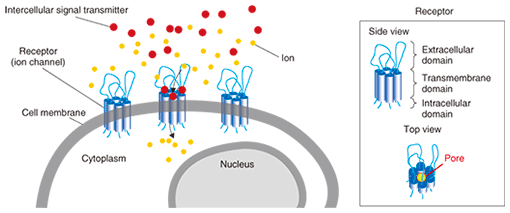
2. AFM observation of ATP receptorThe ATP receptor is activated by binding with ATP, and extracellular cations including Ca2+ and Na+ flow into the intracellular space. A single ATP receptor (i.e., a subunit of a functional ion channel (ATP receptor channel)) has three domains: the large extracellular domain, two cylindrical transmembrane domains, and the N- and C-terminal*1 intracellular domains (Fig. 2, left). In the extracellular domain, six β-strands*2 form interlinking hydrogen bonds, thereby forming a large β-sheet structure [3]. The target of this study was the P2X4 receptor, which is a type of ionotropic ATP receptor that is expressed throughout the central nervous system including the brain and spinal cord. The P2X4 receptor expressed in the microglia of the spinal cord has recently been reported to contribute to the induction of neuropathic pain, a severe pain sensation on which narcotic analgesics have no effect [4]. On the basis of that report, the P2X4 receptor has attracted attention as an important therapeutic target as regards neuropathic pain.
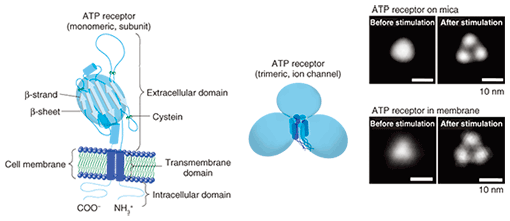
Purified ATP receptor proteins were adsorbed on freshly cleaved mica and observed by AFM. In a non-stimulated condition, the ATP receptor channel exhibited circular features and a trimeric topology after stimulation with ATP (Fig. 2, right). Before ATP activation, the distance between subunits seemed to be smaller than the spatial resolution of our AFM system. These two images were obtained from two different samples treated with and without ATP, respectively, so we cannot say that this structural difference actually derives from the conformational changes in ATP in the ATP receptor channel. To clarify this, we used a recently developed AFM technique called fast-scanning AFM [6] and performed time-lapse imaging of the conformational changes in the ATP receptor channel. Although the ATP receptor channel was circular before stimulation, after stimulation by ATP, it immediately took on a trimer structure (Fig. 3). With prolonged ATP stimulation, the three regions moved farther apart (disengaged). Even though fast-scanning AFM clearly demonstrated that the ATP receptor channel changed its conformation by binding with ATP, it is not clear whether this conformational change is related to a physiological function.
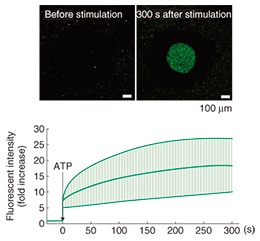
We then estimated the function of a purified ATP receptor channel. A comparison of the predicted structure and the AFM images of the ATP receptor (Fig. 2) shows that the ATP receptor appears to stand upward. From the predicted structure, one would expect the ATP receptor subunit to have six β-strands in its extracellular part, which would bind together to form one β-sheet. This structure should be observed as one large domain by AFM. By contrast, the transmembrane domains are α-helix structures and the intracellular domains are fiber-like structures. In addition to this, since ATP binds with only the extracellular domain of the ATP receptor, if the receptor responds to ATP, the structure observed by AFM should be the extracellular domain of the ATP receptor. We found that the AFM image of the ATP receptor when it was inserted into the lipid membrane was similar to the image of the ATP receptor on mica. When the ATP receptor is inserted into a lipid membrane, it aligns vertically. If the ATP receptor is inserted upwardly into the membrane, but not downwardly, it can bind with ATP and exhibits a structural change. Furthermore, if the ATP receptor is inserted downwardly, it cannot be activated by ATP and shows no Ca2+ permeability. The function of the ATP receptor channel can be estimated by using ion indicators. In our study, we used Ca2+ indicator to detect the Ca2+ flow though the ATP receptor channel and we found that the ATP receptor showed significant Ca2+ permeability after ATP stimulation (Fig. 4). We fabricated 500-¦Ìm diameter wells, filled them with Ca2+ indicator, and then covered them with lipid bilayers. ATP receptor channels were reconstituted in the lipid bilayer, and Ca2+ was added to the buffer outside the wells. The fluorescent intensity of the wells increased immediately after ATP addition (Fig. 4). This result means that ATP binds to the ATP receptor channel and that the ATP receptor channel opens its transmembrane pore. Consequently, calcium ions flow into the well from the space outside it.
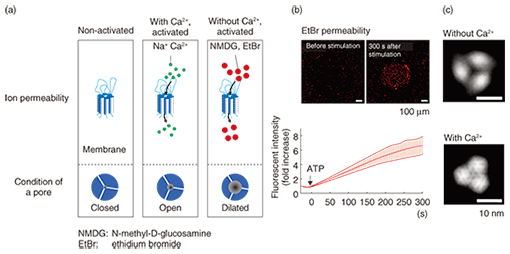
3. Pore dilationIt is known that after its activation, the ATP receptor forms large pores that are different from normal pores. This phenomenon, discovered by Cockroft and Gomperts in 1979 [7], is called pore dilation. The normally opened pore of the ATP receptor channel allows cations including Na+ and Ca2+ to permeate the cell. Of the ATP receptors, P2X2, P2X4, and P2X7 receptors are known to form large pores, but it is unclear whether only the ATP receptor exhibits pore dilation. Direct evidence has been awaited for three decades since the discovery of this phenomenon. The ATP receptor exhibited further disengagement of its subunits after its conformational change into a trimer (Fig. 3). This structure strongly indicates pore dilation, which is schematically shown in Fig. 5(a). To confirm this, we performed a functional analysis of the ATP receptor. The P2X4 receptor is known to exhibit pore dilation only when the extracellular calcium level is low [8]. Therefore, we compared two structures and the permeability of the ATP receptor channel under low and high Ca2+ conditions (Fig. 5(a)). Ethidium bromide (EtBr) (size: 1.4 nm × 1.1 nm × 0.52 nm) and N-methyl-D-glucosamine (size: 1.0 nm × 0.76 nm × 0.53 nm) can be used to estimate the occurrence of pore dilation [2]. The red fluorescence of EtBr is enhanced after binding with double-strand DNA. When DNA is placed in the well, the permeability of the ATP receptor channel to EtBr can be estimated from the fluorescent intensity of the well. The EtBr flow through the ATP receptor channel increased after ATP addition without Ca2+ (Fig. 5(b)) but not when Ca2+ was present. AFM observation found a pore-dilation-like structure for the ATP receptor channel only in the absence of Ca2+ (Fig. 5(c)). When Ca2+ was present, the ATP receptor channel exhibited a trimeric structure but not a pore-dilation-like one.
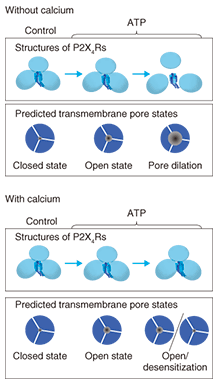
4. New conformational change modelIn the present study, we succeeded in 1) observing a trimeric ATP receptor channel, 2) observing the structural changes in an ATP receptor channel, 3) showing that the structure corresponds to pore dilation, and 4) completing a functional analysis. Although previous studies have found that the intracellular domain exhibits conformational changes in response to activation, no studies on the conformational changes in the extracellular domain have been reported. Using our results, we have established a new conformational change model of the ATP receptor channel (Fig. 6). Without ATP stimulation, the transmembrane pore is closed and the distance between extracellular domains is too short for the detection of individual subunits. After ATP binding, the ATP receptor channel changes its conformation, opens its transmembrane pore, and exhibits a trimeric structure. With Ca2+, no further structural changes are observed. By contrast, without Ca2+, there is further disengagement of subunits and dilation of the transmembrane pore, which becomes permeable to large molecules, including EtBr.
5. Concluding remarksIn this study, we aim to establish basic technologies for the fabrication of new bio-sensing devices that combine biological molecules with nanoscale devices. If we are to introduce biological molecules into devices, it is first essential to understand their biophysical properties. These new bio-sensing devices are expected to lead to a new high-sensitivity information technology. References
|
|||||||||||||||||||||||