|
|
|
|
|
|
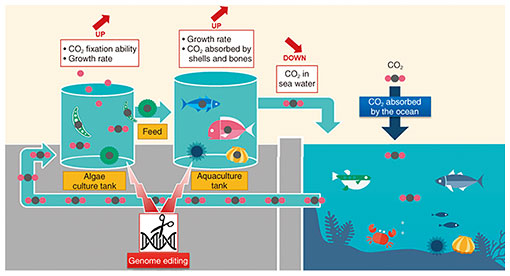
Rising Researchers Vol. 20, No. 5, pp. 7–10, May 2022. https://doi.org/10.53829/ntr202205ri1  Research on Reducing CO2 in the Ocean through the Application of Genome Editing Technology to the Carbon Cycle of Algae, Fish, and ShellfishAbstractAccording to the Intergovernmental Panel on Climate Change (IPCC) Sixth Assessment Report, 57.7% of carbon dioxide (CO2) emissions into the atmosphere are absorbed by forests, and 34.6% is absorbed by the ocean. In this article, we interviewed Distinguished Researcher Dr. Sousuke Imamura about “CO2 reduction technology in the ocean based on application of the genome editing technology.” Keywords: CO2 reduction, genome editing, algae Reducing CO2 in the ocean through genome editing in algae, fish, and shellfish—How is genome editing technology used to reduce the amount of CO2 in the ocean? According to the Intergovernmental Panel on Climate Change (IPCC) Sixth Assessment Report, only 4.8% of total carbon dioxide (CO2) emissions into the atmosphere come from human activities. Of the remaining emissions, soil accounts for 61.3% and the ocean accounts for 33%. On the other hand, up to 57.7% is absorbed by forests, and the ocean also absorbs 34.6%. NTT’s communications services emit CO2 as energy is consumed, so researching and developing technologies that reduce these emissions is vital both for the unimpeded development of the communications business and to reduce our carbon footprint. In addition to CO2 reduction measures such as energy conservation, another feasible approach to absorbing and reducing CO2 emissions is to harness the power of living organisms. Our focus is on the carbon cycle of the ocean, and we’re conducting research into reducing CO2 through the use of genome editing technology. Figure 1 shows an overview of the technology used to reduce CO2 in the ocean by applying genome editing technology to the carbon cycle of algae, fish, and shellfish. When atmospheric CO2 dissolves into the ocean, it is fixed by ocean-dwelling phytoplankton such as algae. The term “fixed” may sound strange, but it refers to the process of carbon fixation, in which inorganic carbon such as CO2 is converted into organic carbon such as sugar and assimilated into the body. The carbon fixed by the algae is then taken in by fish and shellfish that eat the algae as food. This is the carbon cycle of the marine food chain.
We’re trying to apply genome editing technology to key parts of the marine food chain, including algae, fish, and shellfish, which is a world first. As a result, we aim to encourage increased CO2 fixation in algae and accelerate the growth of fish and shellfish, thereby increasing the amount of CO2 absorbed by their bones and shells. As such, this research is aimed at synergistically increasing the total amount of carbon held in the carbon cycle by algae, fish, and shellfish, thereby reducing the amount of CO2 present in seawater. As the concentration of CO2 in seawater decreases, the amount of CO2 dissolved from the atmosphere increases accordingly, thus reducing the amount of CO2 in the atmosphere. We’ve started a proof of concept with Regional Fish Institute, Ltd. (RFI), in which NTT will work on research and development of genome editing technologies that increase the amount of CO2 fixation in algae, and RFI will work on research and development of genome editing technologies that increase the amount of carbon stored in the bodies of fish and shellfish. —What drew your attention to algae? Algae are identifiable as a phytoplankton that contribute to CO2 fixation in the ocean, so they seemed like a natural area to focus on. They’ve been around for some three billion years, all the while producing oxygen through photosynthesis. No other organisms have a history comparable to that. We’re breathing now thanks to the oxygen they’ve stored in the atmosphere, and the evolution of plants we’re familiar with today can be traced back to algae. It’s no exaggeration to say that algae have shaped the global environment as we know it. I felt algae would be a good route to understanding plants, so I’ve been studying them since my undergrad days. In addition, since many of algae are single-cell organisms, they are easy subjects for genome editing. —What exactly is genome editing technology? The term “genome editing” is often confused with genetic modification. It’s common to come across disclaimers about genetic modification on food packaging. For example, in Japan, you see messages like “does not contain genetically modified soybeans” on natto (fermented soybean) packages. Genetic modification involves taking genes derived from one thing and inserting them into another, like if you added photosynthesis genes from algae to the human body. Genome editing technology, meanwhile, is about altering genetic sequences, or subtly editing the order of genes, to put it another way. It’s considered relatively safe compared to genetic modification, because it doesn’t involve inserting genes derived from an external organism into the body of another—it’s just a case of changing gene sequences that are already there. It’s the same basic principle as selective breeding of crops. That said, it is vital that we carefully assess the environmental impact of genome-edited organisms and whether or not it could lead to any damage to biodiversity. That’s why we’re conducting our proof of concept in a confined space through land-based aquaculture (Fig. 1). Our goal over the next five years is to put our first set of algae and sea creatures to practical use—Can you tell us about where you’re currently at with this research? Right now we’re conducting research on three fronts. The first is research on feeding. We’ve started actually testing feeding the cultivated algae to the fish and shellfish, but as things stand, it seems that they have very particular preferences and won’t really eat the algae. So we’re looking into which types of algae they’ll ingest more efficiently, and at what concentrations. Our second area of research is genome editing. When editing genomes, you need to determine which sequences of which genes you want to edit. We’re currently investigating the best conditions for editing genomes in order to produce the desired effect. Our third point of research is reducing unwanted side effects caused by genome editing. As I mentioned earlier, algal mechanisms have been accumulated and refined over three billion years. If human beings start meddling with those mechanisms, there are bound to be consequences. For example, increasing CO2 absorption or fixation ability may have a negative impact on growth. So we’re researching the best ways of mitigating such trade-offs. —What are your plans for future research? The ocean is home to a remarkably wide variety of species. I began by outlining the percentages of CO2 emissions and absorption, but these are just calculations at the end of the day. We can only make rough estimates derived from the assumption that all organisms would function in similar ways, based on existing data from a selection of algae and other species. We hope that by experimenting with land-based aquaculture in a confined space, we’ll have a platform to generate quantitative data on the levels of CO2 fixation in algae, to what extent the carbon is passed to fish and shellfish, and the overall effect on CO2 levels. This approach will be an important point of focus for future practical applications. Our goal over the next five years is to determine the best combination of Alga A and Fish or Shellfish B, to evaluate the effectiveness and safety of this CO2 reduction method, and to put our findings to practical use. We should then be able to explore how this technology will be used out in the actual ocean environment, not just within a confined space. So far, I’ve talked about reducing CO2 from an environmental perspective, but another important consideration is food. RFI has already successfully edited genomes to develop fish that could compensate for food shortages, such as red sea bream with more edible parts and tiger pufferfish that grow at an accelerated pace. By feeding algae that efficiently fix CO2 in the ocean to genome-edited fish and shellfish, this technology should be capable of both reducing CO2 levels and producing food at the same time. That said, genome editing is still a new and relatively untried technology, so it’s essential to provide consumers with accurate information in order to dispel any concerns they might have about genome-edited food. In terms of shellfish, we’re also conducting research into Akoya pearl oysters. Even people who are averse to eating genome-edited food may find it easier to accept decorative objects such as the pearls produced by these oysters. Of course, our main objective is to increase CO2 fixation, but there will likely be a growing need to develop research like this that adds value to fish and shellfish in the future. We’re also considering how to apply this technology to soil. Soil accounts for the largest proportion of CO2 emissions, and crucially, it’s the source of nutrition for plants. Any reduction in the amount of CO2 generated from soil is important, no matter how small. Although there are many differences between soil and ocean, microorganisms play a huge role in both, so I have high hopes for some breakthrough findings in this area. —Is there anything you would like to say to students, young researchers, and future business partners? In order to accelerate the research I’ve described today and develop it into a more versatile technology, we’re looking to work together with data scientists who can help us digitize and analyze data relating to genome editing, algae cultivation and feeding, as well as collaborating with people who specialize in entire ecosystems. At NTT Space Environment and Energy Laboratories, we conduct research in an extensive range of fields. In addition to research like ours that explores and pursues a deeper understanding of life, we conduct research into the fundamentals of communications, networks and the engineering that supports these, as well as areas like energy and social sciences. We are working on new and challenging themes that have never been tackled before. I joined this research lab in March 2021 straight from the world of academia, and am excited to be working on such large-scale and well-balanced projects, from research through development. At present, there remains a great deal we don’t know about various biological mechanisms. In this field alone, I expect many new ideas to come to the fore, which could then develop into new areas of research as our understanding grows. NTT Space Environment and Energy Laboratories is an environment in which you can challenge yourself to try new things. If you’re an innately curious person, we’d love to have you on board. ■Interviewee profileBefore joining NTT in 2021, Sousuke Imamura completed a doctoral course in biotechnology at the United Graduate School of Agricultural Science at the Tokyo University of Agriculture and Technology, became a research fellow at the University of Tokyo, gained a Research Fellowship PD from the Japan Society for Promotion of Science at the University of Tokyo, became an Assistant Professor in the Department of Life Sciences at the Chuo University Faculty of Science and Engineering, and became an Associate Professor at the Laboratory for Chemistry and Life Science in the Institute of Innovative Research at Tokyo Institute of Technology. He is a member of the Sustainable Systems Group at NTT Space Environment and Energy Laboratories, as well as a lecturer at Meiji University’s School of Agriculture and a visiting professor at the Laboratory for Chemistry and Life Science in the Institute of Innovative Research at Tokyo Institute of Technology. His specialty is plant molecular biology. |
|