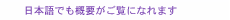|
|||||||||||||
|
|
|||||||||||||
|
Feature Articles: Recent Updates on Bio-soft Materials Research Vol. 22, No. 5, pp. 31–38, May 2024. https://doi.org/10.53829/ntr202405fa3 Brain-on-a-chip Model Using Deformable Graphene-based Electrode ArrayAbstractSince the brain transmits electrical signals between neurons in three-dimensional (3D) space, we developed a 3D deformable electrode array to measure electrical signals from a 3D network of cultured neurons using a graphene self-folding technique. By recording the electrical signals spatiotemporally from the electrode array, we demonstrated that the reconstruction of a 3D neuronal network on the electrode array (brain-on-a-chip model) enables us to investigate how neurons communicate with each other and helps us understand how the structure of the neuronal network affects the neuron’s communication. Keywords: organ-on-a-chip, neural interface, neuronal network 1. Brain-on-a-chip modelThe function of living tissues is highly dependent on their structure. The tubular structure of blood vessels and intestines allows for the flow of substances, and the fibrous structure of skeletal muscles generates traction force. The brain also has structural features, including three-dimensionality and modularity. For example, the cerebrum is divided into motor, somatosensory, and visual cortices, which have different roles. Within each area, neurons are not uniformly distributed but maintained in dense clusters of cells and weakly connected clusters of cells. These weak connections between cell clusters with specific roles are called modular structures, and the brain functions require a balance of connectivity between modules that work synchronously or individually. We investigated how these brain structural features affect neuronal synchronization at the cellular level by growing cultured neurons on an electrode substrate. Cultured neurons obtained from brain tissue can still generate electrical signals. Furthermore, cells growing on the substrate spontaneously form a neuronal network so that the communication between cells can be observed from electrodes placed at multiple points. However, cells on a flat substrate grow uniformly and in a flat plane in a structure completely different from that of the brain. Therefore, it is necessary to artificially manipulate the structure by constructing scaffolds for cultured neurons to grow and by localizing the cells to reproduce the brain structural features, including three-dimensionality or modularity. We propose a strategy in which the electrodes are used as three-dimensional (3D) scaffolds to use the electrodes to measure the electrical signals of the cells and arrange the cells to form the desired 3D structure. In this article, we introduce a 3D deformation technique of electrodes and the growth of cells in 3D structures using the electrodes as scaffolds. We then reproduce both 3D and modular neuronal networks on an electrode array by connecting 3D cell clusters (Fig. 1). The development of a brain-mimicking neuronal network on the chip is called a brain-on-a-chip model. After introducing the results of recorded neural signals from the brain model, we provide an overview of further development of the technology.
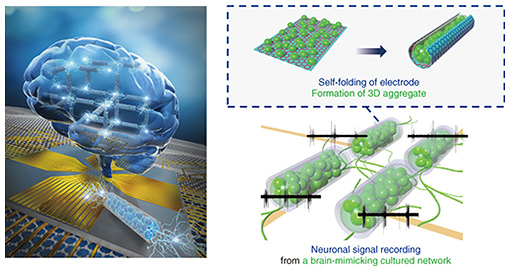
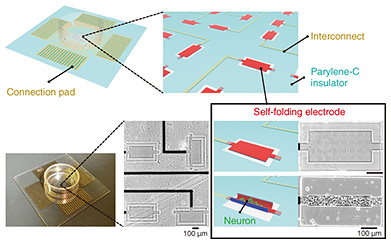
2. Cell encapsulation within the 3D deformable electrodeTo use electrodes as 3D cell scaffolds, we developed a method of folding electrodes three-dimensionally and encapsulating cells within them [1–3]. The procedure of this method is shown at the top of Fig. 2. The process of growing cultured cells three-dimensionally has been actively studied in tissue engineering and organ-on-a-chip technology. However, it is challenging to apply 3D culture methods to recording techniques with a flat electrode substrate (microelectrode array) because poor contact between the flat electrode and cultured 3D tissue decreasing signal amplitude. This is one of the fundamental reasons for the difficulty of developing sensing technology in organ-on-a-chip. Therefore, electrodes must have a 3D curved structure to be compliant with the surface of the 3D tissue. Current planar electrodes can be fabricated by photolithography with a spatial resolution corresponding to the size of cells (10–100 μm scale). The processing accuracy of photolithography supports biosensing technology for measuring cellular signals. However, it is challenging to form 3D electrodes with precise geometries due to the technical difficulty of manually bending precisely fabricated electrodes.
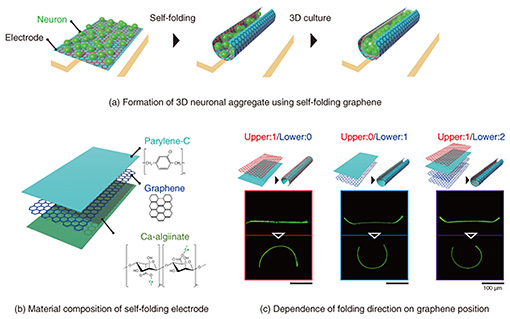
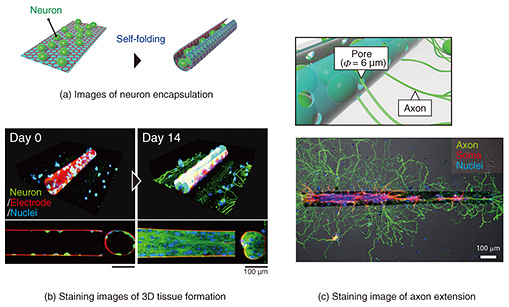
We incorporated a technique called thin-film self-assembly into an electrode array substrate to bend a precisely fabricated electrode. In a bilayer thin film of two different materials, the film folds when one of the layers is subjected to shrinkage or stretching forces. Self-assembly is a method of inducing spontaneous folding of a thin film when detached from a substrate by preparing a bilayer thin film on a substrate and applying internal stress to one of the layers in advance. Typical examples of deformation include transforming a thin rectangular film into a cylindrical shape and transforming a dice net into a cube shape. We developed a self-folding thin film composed of biocompatible materials [4]. By using these techniques and knowledge, we have further explored materials that can be used as electrodes. As a material for a self-folding electrode, we selected graphene consisting of one layer of carbon atoms because it is highly biocompatible, electroconductive, and flexible enough to be bent (Fig. 2(b)). Graphene has also been reported as an electrode material of brain-implantable devices. After screening several candidate materials for the self-folding thin film, we found that a polymer material called parylene-C can induce self-folding by attaching graphene to a parylene-C sheet [2, 3, 5]. As shown in Fig. 2(c), the graphene/parylene-C bilayer is flat while attached to the substrate, but once it is removed from the substrate, it folds into a cylindrical shape within a few seconds. The critical point is the high adhesion of the graphene to perylene-C. Because of the high adhesion, graphene does not slip from the perylene-C sheet during the folding, and the residual stress generated inside the film is the driving force for the folding. Parylene-C is another material widely used for the insulating layer of neural electronic devices, and its excellent biocompatibility and transparency also make it suitable for microscopic cell observation. Material candidates for a sacrificial layer, which supports the bilayer thin film on the substrate, were also screened. Calcium alginate gel was selected as the material for the sacrificial layer because it does not affect the survival of cultured cells during dissolution. The calcium alginate gel can be dissolved with a reagent that removes calcium ions (Ca2+) without affecting the survival of cultured cells. After adding the Ca2+-chelating reagent, the thin film immediately detaches from the substrate and completes the self-folding. After stacking three layers of thin films containing calcium alginate gel, parylene-C, and graphene, the tri-layers are etched with oxygen plasma through a photolithographically patterned mask, allowing the thin films to be shaped with micron-meter-scale precision. Rectangular films from 100 to 1000 µm are typically formed to obtain a cylindrical shape with a curvature radius of 10–200 μm after self-folding. The curvature radius depends on the thickness of the parylene-C film. For the bilayer of graphene and parylene-C film, the graphene becomes the outer surface of the cylindrical structure (Fig. 2(c)). When cultured cells are wrapped, they can only contact the insulating parylene-C thin film. We fabricated a sandwich structure of graphene and parylene-C thin film, as shown on the right side of Fig. 2(c), and found that this tri-layer film can also spontaneously fold. Thus, we developed a self-folding electrode with an inner conductive graphene layer [6]. Since graphene is not only biocompatible but also highly transparent, cells can be fluorescently stained and observed under a microscope to visualize the process of cell growth inside the folded electrode and the formation of 3D neuronal aggregate (Fig. 3). A culture medium containing neurons obtained from the rat hippocampus is dropped onto an electrode. Self-folding is then induced, resulting in the encapsulation of neurons in the folded electrode (Fig. 3(a)). Figure 3(b) shows neurons are attached sparsely inside the folded electrode on the first day of culture. They grow and elongate their process, increasing the tissue volume formed by cell aggregation. After 1–2 weeks of culture, the tissue volume increases to fill almost the entire inside of the tube, forming a 3D neuronal aggregate along the shape of the tube. After 3–4 weeks of culture, most neurons are still alive, and the 3D tissue is maintained inside the cylindrical electrode. The electrode structure is modified to maintain the culture for such a long period. If the culture medium is simply decreased, cells cannot survive due to a lack of nutrients and oxygen. Long-term culture of 3D tissue requires a continuous supply of culture medium to the inside of the tissue. It is challenging to supply culture medium from the outside culture using the folded structure. Therefore, as shown in Fig. 3(c), we incorporated pores into the electrode surface to improve the material permeability [1, 2]. By using the resolution of photolithography to form tiny pores (3–6 μm), the permeability of nutrients and oxygen can be improved while keeping the neurons confined inside the tube.
The role of the pores is not only to improve permeability. Pores also function as a pathway for neurons to connect structurally to the outside of the tube. Neurons connect to other neurons by extending thin projections called axons to form a neuronal network. To achieve the modular structure described at the beginning of this article, it is essential to provide tiny pores so that 3D neuronal aggregates can be connected through axons. The bottom of Fig. 3(c) shows a stained image of an axon (green) extending outside the tube through tiny pores. The soma (cell body; red) remains inside the tube, and only the axons can be extended outside the tube and connected to other 3D neuronal aggregates. This method solves the technical problem of structural mismatch between 3D neuronal aggregates and electrodes and enables the connection of 3D neuronal aggregates by providing tiny pores for axons to pass through to form a modular neuronal network. 3. Brain-on-a-chip model using self-folding electrode arrayIn addition to three-dimensionality, the modular structure is another feature of the brain. We developed an electrode array chip in which multiple self-folding electrodes are aligned to form the modular structure by connecting multiple 3D culture tissues encapsulated within folded electrodes [6]. As shown in Fig. 4, a neuronal network is formed by arranging self-folding electrodes in an 8 × 8 multi-array of substrates and forming 3D neuronal aggregates inside each electrode, with each 3D neuronal aggregate as a module. Neurons spontaneously extend their axons, and the 3D neuronal aggregates are connected. Weak connections are formed between the neuronal aggregates compared with the dense connections between the neurons in the 3D neuronal aggregates, forming a modular structure with distinctly solid and weak connections. By independently wiring each electrode to an external recording device (Fig. 4), neural signals exhibited from cultured 3D tissues can be recorded separately. By analyzing the synchronization and time delay of the recorded neural signals, it is possible to investigate how the multiple 3D neuronal aggregates are connected and how the neuronal network is formed. This engineered neuronal network on the chip device functions to reproduce the structural features of the brain while electrically visualizing the interactions within the neuronal network. We conducted experiments for testing two points: long-term measurement performance and the effect of the structural features of the network on the synchronization of neural firing.
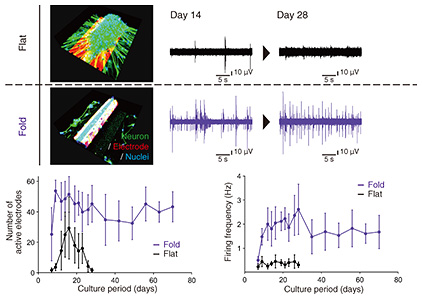
We investigated long-term measurement performance by comparing the recorded signals between the self-folded 3D electrode and the existing flat electrode. The deformation of the electrode can solve the structural mismatch between the flat electrode and 3D neuronal aggregate. We evaluated how long neural signals could be stably recorded from individual 3D neuronal aggregates. Figure 5 summarizes the recording results. As a comparison, the recorded signal decreased and disappeared as the culturing days progressed when a 3D neuronal aggregate was formed on a conventional planar electrode. In addition to the structural mismatch, the cause of decreased signals is the ability of cultured cells to migrate. As shown at the top of Fig. 5, the movement of the 3D neuronal aggregate from the flat electrode decreased the contact area, and the recorded signal gradually weakened. However, the three-dimensionally deformed tubular electrode maintained good contact between the tissue and electrode because the 3D neuronal aggregate was fixed inside the tubular electrode. The number of electrodes that could be recorded and the recorded neural firing frequency as a function of the culturing period is shown at the bottom of Fig. 5.
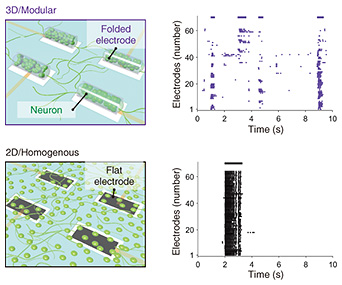
We then examined the relationship between the structure of the neuronal network and synchronization of neural firing. In neuronal networks, neurons fire synchronously and generate electrical signals. In modular structures, neurons are tightly connected to each other within a module that is a 3D neuronal aggregate and tend to fire synchronously, while neurons are loosely connected between different modules and are relatively less synchronized. To investigate the effect of such structural features on synchronization, we recorded spontaneous activity from networks with modular and 3D structures (3D/modular networks). We compared the recorded signals with those recorded from neuronal networks in which cells are uniformly grown on the planar substrate (2D/uniform networks). Figure 6 shows the results of spontaneous activity recorded at each electrode as a plot of the firing time at each electrode for visualizing the synchronization between the 3D neuronal aggregates. In the 2D/uniform network, the firing pattern is dominated by the repetition of synchronized firing due to the strong connection.
In the 3D/modular network, a variety of firing patterns in which several 3D neuronal aggregates synchronize with each other was increased. The firing patterns included neural firings that do not participate in the synchronization. This result indicates various firing patterns in which several modules fire synchronously while some fire asynchronously. In the brain, it has been theoretically shown that such a modular structure that generates diverse firing patterns with a mixture of synchronous and asynchronous firing is advantageous for information processing. In this study, we experimentally demonstrated that the balance between synchronous and asynchronous firing fluctuates by reproducing modular and 3D structures in the cell culture model. The changes in the synchronous firing can be evaluated for an extended period. For example, after 7–9 days of culture, when axons bridge the 3D neuronal aggregates, asynchronous firing gradually becomes synchronous. After 2–3 weeks of culture, the overall synchronous firing becomes dominant, and the interval between repeated synchronous firings becomes shorter. The electrophysiological recordings help us better understand the formation of neuronal networks related to the brain’s developmental process. Thus, the brain-on-a-chip model constructed in this study enables us to investigate brain systems related to the structure and maturation of neuronal networks from their firing patterns by changing the culture time and shape parameters such as electrode size and spacing. 4. Summary and future perspectiveIn this article, we demonstrated modeling neuronal networks in the brain by culturing cells on the electrode array while reproducing 3D and modular structures characteristic of the brain. Even single neurons have more exciting properties than other cells, but their collective behavior changes depending on the culture conditions, including the structure and materials of the substrate. Although the environment is fundamentally different from that of the actual brain, there is a possibility that new findings related to brain function and development will be revealed by exploring the preserved characteristics of cultured neurons. The closer the structure and properties of the neuronal network are to those of the brain, the more we can uncover relationships between the behavior of the neurons and brain function. In addition to physiological phenomena, this technology also has the potential to be a pathological model. For example, it would be used to construct a neuronal network that simulates autism, which is known to be associated with abnormalities in the structure of neuronal networks. Another disease model could be constructed by mixing neurons from induced pluripotent stem (iPS) cells derived from patients with neurological diseases. This model can be used to investigate drug responses to neuronal networks with brain-like functions without using the actual brain, contributing to medical research and drug discovery by reducing the number of experimental animals and accelerating drug testing. The performance of this model will be improved by increasing the number of electrodes, adding multimode stimulations (light, heat, vibration, etc.), and increasing the variety of 3D shapes of the electrodes. Targeting events related to brain function and pathology will enable us to develop advanced brain-on-a-chip models that better reflect physiological phenomena and diseases. References
|
|||||||||||||