|
|
|
|
|
|
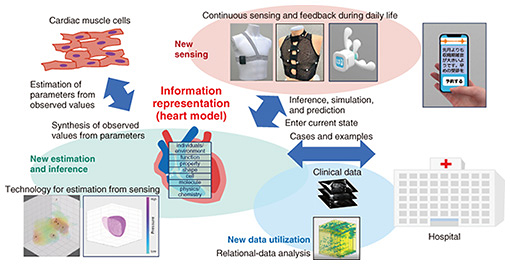
Front-line Researchers Vol. 22, No. 9, pp. 6–10, Sept. 2024. https://doi.org/10.53829/ntr202409fr1  Creating Bio-digital Twins by Using Crossmodal Representation LearningAbstractAnnounced in November 2020, NTT’s Medical and Health Vision states that the company will strive to put biological simulations using bio-digital twins (BDTs) into practical use to make effective use of medical resources, alleviate physical constraints of medical resources, provide continuous care from prevention to post-treatment, and provide precise care personalized for each individual. A BDT is a digital representation of a living organism, and the key to creating a BDT is how to express (quantify or symbolize) the functions of the living organism—and the physical and chemical mechanisms behind them—as digital information. We interviewed NTT Fellow Kunio Kashino of NTT Communication Science Laboratories, who has taken the challenge of creating a BDT, about the use of artificial intelligence in the biomedical field, stimulation and awareness in collaborative research across different disciplines, and his thoughts on the importance of striking a balance between what should be done and how it should be done. Keywords: representation learning, crossmodal approach, bio-digital twin Generating new knowledge through crossmodal approach and applying it to the biomedical field—Can you tell us about the research you are currently conducting? I am currently focusing on representation learning and basic research on biomedical informatics. I have been researching sound and image/video recognition and retrieval for a long time. The key point of this research is how to represent media information accurately and efficiently as digital information, which involves representation learning, my first focus. I have recently expanded my research beyond accurate and efficient information representation to include information representation that can uncover hidden structures in data and support the discovery of new knowledge. My second focus, basic research on biomedical informatics, involves the application of representation learning in the medical and health fields. Generative artificial intelligence (AI) based on large language models has been attracting attention. Generative AI also operates on the basis of the information representation (a digital representation of information) of something and generates output appropriate to a given condition (e.g., a question in sentence form) while referring to the information representation. How to represent information is learned from large amounts of data. The power of large amounts of data is so overwhelming that it is beyond even the imagination of experts, and AI systems are being created that produce useful outputs, such as text, images, and video, that would have been unthinkable not long ago. However, some aspects of these systems are not fully understood; for example, it is not known why a system behaves in the way that it does or how specific information is represented within a system. The above-mentioned research on representation learning acknowledges this problem, and it can be said that it aims to achieve both the advantages of learning based on large amounts of data and the transparency of information representation. Therefore, if this research is successful, it should provide answers to questions such as how to optimize the size of AI systems and ensure the reliability of their output. My research topics on representation learning include elucidation, analysis, construction, optimization, and application of information representation. My goal is to establish a method of configuring an optimized information representation with a clear mechanism of action. To achieve this goal, I’m now focusing on extracting and using the relationships between different pieces of information. Let’s consider the problem of image recognition as an example. If image data are the only reference, the usual approach is to prepare a large amount of training data consisting of pairs of images labelled with the names of the objects depicted in each image and train the AI with that data. Although this approach is very powerful when it works, it cannot always easily create suitable training data and is not suitable when the way objects are named changes over time. However, when an image and its associated audio information are available, such as online videos, television programs, and everyday scenes, it is unnecessary to manually prepare training data; instead, it is possible to use the relationship between the image and audio as a clue to identify the image and even determine the semantic relationship (closeness of meaning) between the objects shown in the image. Our experiments have shown that if an AI system is given hundreds of hours of recorded sumo-wrestling broadcasts without any prior knowledge, it can identify frequently occurring (e.g., top 10) winning moves from only image and audio information with a reasonable degree of accuracy. As in this example, it is often the case that something is difficult to understand by referencing just one type of data, but it becomes clear by referencing multiple types of data. The types of information are called “modalities,” and I believe that one of the keys to representation learning in the future will be to study these modalities and analyze their relationships with each other. We call this the “crossmodal approach.” It is now possible to collect a large amount of various types of information. In the field of medicine and biology, recent research and technological advances are gradually making it possible to analyze and cross-check large amounts of different types of information, such as genetic information, the cellular basis of behavior, and clinical test results. The increased deployment of the Innovative Optical and Wireless Network (IOWN) will further strengthen these advances. By clarifying the hidden relationships between information collected in this crossmodal manner and reflecting them in inferences and simulations, I believe that—in the not-too-distant future—it will be possible to predict the state of a person’s health several years into the future and estimate the efficacy and side effects of drugs and treatment methods to a certain extent. I also believe that through this type of crossmodal research, we can create AI that generates new knowledge that humans have not been aware of. AI may also be able to demonstrate its creativity as a useful partner to humans in scientific and technological research. —Applying crossmodal approach to biological information leads to bio-digital twins, right? Yes. Modeling of living organisms has been around for a long time, and you could even say that the history of medicine and biology is the history of modeling. Most such modeling was based on individual experiments and insights of experts. Living organisms are, however, complex subjects, so creating detailed models by hand is naturally limited. To harness the potential power of large volumes of or diverse data, technology that can automatically learn information representations is therefore important. In November 2020, NTT announced its Medical and Health Vision to contribute to the effective use of medical resources, alleviation of physical constraints of medical resources, provision of continuous care from prevention to post-treatment, and provision of precise care personalized for each individual. The core concept is simulating living organisms using bio-digital twins (BDTs). Our AI telestethoscope introduced in the previous interview (August 2021 issue) is one of the sensing tools used for actualizing this concept. Later, the Biomedical Informatics Research Group was newly established in the Media Information Laboratory of NTT Communication Science Laboratories to support BDTs from the perspective of basic research on informatics, and I was appointed as its leader. Although the basic activities of this group are concerned with creating new basic technologies for machine learning, signal processing, and pattern processing for a wide variety of information, the group is also actively applying these technologies to biological information and developing new socially useful fields of application. We are collaborating with not only the Bio-Medical Informatics Research Center of NTT Basic Research Laboratories and the Alliance Department of the Research and Development Market Strategy Division at NTT but also universities and hospitals that have unique strengths. Let me introduce specific studies undertaken by my research team. We are collaborating with Sakakibara Heart Institute on high-precision simulation of the cardiopulmonary function. Heart disease ranks first or second among causes of death in many countries, and early detection and treatment as well as post-treatment rehabilitation (exercise therapy) are known to be particularly effective. Exercise therapy, in particular, can dramatically improve five-year survival rates by having patients exercise at an intensity appropriate to the individual. However, the question is how to set the appropriate intensity of the exercise. Too little exercise will be ineffective; too much exercise could be counterproductive or even dangerous. When exercise is prescribed, a test called cardiopulmonary exercise testing (CPX) is therefore used to set the exercise intensity. However, CPX is not widely used because it requires the subject to exercise close to their limit. Given that situation, using data from Sakakibara Heart Institute, which has conducted the largest number of CPX in Japan, we used machine learning to create a model that can estimate CPX results from physical findings without actually conducting a CPX. We hope that this model will enable more people across the country and around the world to receive appropriate exercise therapy. We are currently preparing to practically apply the model in the near future through various verifications and trials. We are also working with the Premium Research Institute for Human Metaverse Medicine (PRIMe) at Osaka University on performance measurement and modeling of cardiac muscle cells by using induced pluripotent stem (iPS) cells. As many readers know, iPS cells have the ability to differentiate into cells of any type of tissues and organs by manipulating cells taken from human skin, blood, and other sources in a specific manner and culturing them. Osaka University is using this capability of iPS cells to study disease models using artificial cardiac muscle tissue. In other words, this study aims to explain the mechanism of heart disease and develop treatment methods by culturing cells taken from heart disease patients with genetic factors, growing the cells into cardiac muscle tissue, and measuring their properties as cardiac muscle, such as contractility and diastolic force. This is a physical disease model, but we are working on digitizing the model in collaboration with Osaka University while measuring contraction and expansion forces. Among the many advantages of digital modeling, I believe the most important is the possibility of creating a model of the heart (i.e., an organ) by synthesizing cardiac muscle tissue in digital space. The model connects the numerous pieces of available information about the heart with microscopic biological information about cells and genes (Fig. 1). It is currently difficult to physically construct organs with complex structures because the cell mass that can be produced in culture is small compared with an organ, i.e., a few millimeters to a few centimeters in diameter. In digital space, however, it may be possible to synthesize cell masses together and estimate their performance as an organ under certain conditions and assumptions. I believe this will be a significant step forward in improving treatment options.
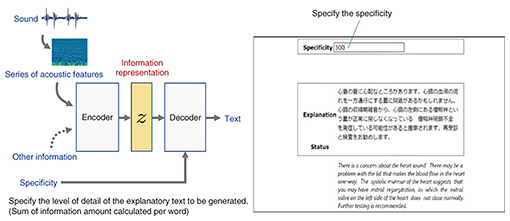
Regarding research on multi-channel, multi-modal biomeasurement using the AI telestethoscope, which uses AI to analyze data collected from electrocardiogram electrodes, microphones, pressure sensors, acceleration sensors, etc. as well as sounds (such as heart and internal sounds) to infer physical conditions remotely (Fig. 2), we are aiming to create new use cases of the AI telestethoscope and improve its accuracy by inputting the measured data into a crossmodal encoder/decoder and displaying explanatory text on the basis of the acquired information representation. In a joint research project with Kitasato University Hospital, we are currently verifying the practicality of a stethoscope-type sensor device that is placed on the chest. The findings that we obtained have recently been published in a medical journal.
I am also pursuing research on the fundamentals of machine learning. During the cell-differentiation process, for example, cells obtain different functions in accordance with the biological tissue, and it is important to analyze, estimate, and model the original state of the cells (before cell-differentiation process) from observations at certain points in time. From a machine-learning perspective, the challenge is how to construct a model with high probability in a situation with many uncertainties. We are developing and verifying such a method. Overcoming barriers to collaborative research across different disciplines—While the work you have described is basic research on technologies. What are the challenges toward practical use of these technologies? First, from the perspective of research, I think the fact that many members of my research team—including me—were not experts in medicine or biology was a challenge. Collaboration across disciplines requires a certain amount of basic knowledge of disciplines different from us, and since the language and behavioral styles are often completely different, imagination and a willingness to compromise are required for successful collaboration. Although my team members still face many challenges, they also enjoy surprises and new discoveries. From the perspective of practical application, when we are dealing with living body and medical care, we must ensure reliability in accordance with established rules, which is an area outside our expertise. Fortunately, we have the cooperation of many people in this area, and I am grateful that we are gradually making progress. By understanding each other’s differences in common practice and culture, important things that people have in common will become apparent—What do you keep in mind as a researcher? It may sound surprising, but I try to be very conscious of social perspectives in my work, which is centered on basic research. NTT Group is becoming an ever more global corporate entity, but I have found that most of my contact with the world has been through participation in international conferences and meetings and other contacts within the research community. Recently, however, as I have come into contact with the field of biological information, I have come to think more about the lives of ordinary people living in various environments around the world. Another point I try to keep in mind is the balance between what should be done and how it should be done. Which of these two questions is more important depends on the situation, but as far as the creation of new technology is concerned, I believe that both are important. I believe that the dynamism of a situation in which new methodologies increase what can be done and the pursuit of what should be done accelerates the birth of new methodologies is the driving force for change in the world. To add to my recent impressions, I have had more opportunities to come into contact with medical professionals and been impressed by their sense of mission and sincere attitude toward matters. I feel that by sharing what is important with people from different disciplines and mutually understanding each other’s common practice and cultural differences, important things that people have in common will become apparent. This approach is similar to a mechanism in which essential information is brought to light through crossmodal information processing. —What is your message to future researchers? I would be happy to share with you the challenges and joys of creating something new. Let’s think about and focus on what is important and challenge ourselves without being bound by common practice. ■Interviewee profileKunio Kashino received a Ph.D. in electrical engineering from the University of Tokyo in 1995 and joined NTT Basic Research Laboratories the same year. He was a visiting scholar in Cambridge University in 2002. His research interests include audio and crossmodal information processing, media search, and biomedical informatics. He is a fellow of the Institute of Electronics, Information and Communication Engineers (IEICE), and a member of the Institute of Electrical and Electronics Engineers (IEEE), the Association for Computing Machinery, Information Processing Society of Japan, the Japanese Society for Artificial Intelligence, and the Acoustic Society of Japan. He received IEEE Transactions on Multimedia Paper Award in 2004, Maejima Hisoka Award in 2010, Kenjiro Takayanagi Achievement Award in 2016, IEICE Achievement Award in 2017, and the Commendation for Science and Technology by the Minister of Education, Culture, Sports, Science and Technology in 2019. |
|